Rare Disease Day is fast approaching, with a programme of activity being lined up, through international initiatives and country-specific programmes, in the UK for example to highlight challenges, raise awareness and generate change for the 300 million people worldwide living with one or more of the roughly 8,000 known rare diseases, as well as their families and carers.
A substantial portion of people with rare diseases face long delays in getting an accurate diagnosis; symptoms may be non-specific, overlap with more common conditions, or vary significantly between patients. This leads to a high rate of misdiagnosis or delayed diagnosis, resulting in a large undiagnosed population, meaning that the total population living with such conditions may be much greater than current estimates, as illustrated in the following graphic:
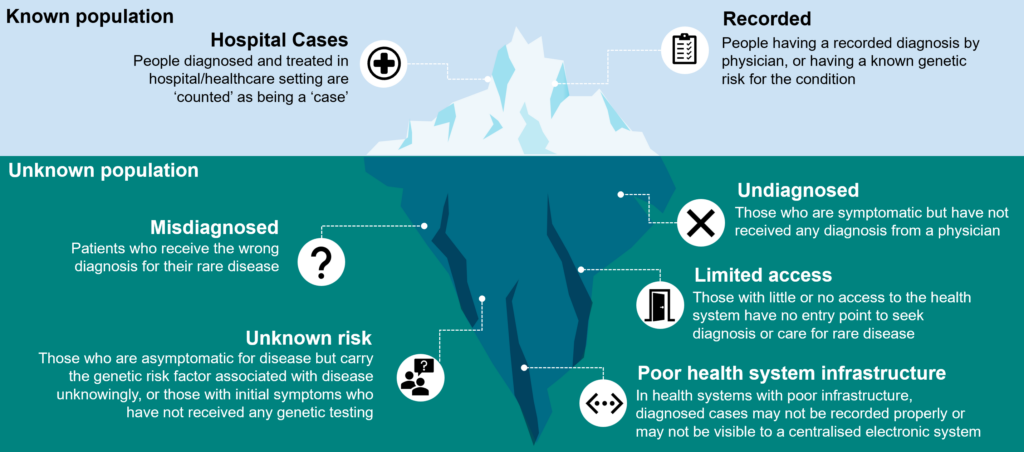
As medical knowledge advances, new rare diseases are identified, and existing diseases are better understood, which can change the number of undiagnosed individuals.
There is also no universally agreed definition of what constitutes a rare disease. Different countries have different thresholds for considering a disease rare.
- For example, in the USA a disease is considered rare if it affects fewer than 200,000 people across the country at any given time, a definition established by the Orphan Drug Act of 1983, primarily to encourage the development and production of orphan drugs for the treatment of rare diseases.
- Whereas in the EU a disease is considered rare when it affects not more than 1 in 2,000 people within the general population, a definition which is part of the EU’s regulation on orphan medicinal products.
Furthermore, as there are thousands of rare diseases, many of them are poorly understood, and this diversity makes it difficult to gather comprehensive data. Obtaining accurate data is further made more challenging due to the lack of reporting and data collection: many countries do not have specific registries or reporting systems for rare diseases, making it hard to track cases accurately.
As covered in some previous blog posts, at HealthLumen we have been working with large genetic datasets such as gnomAD and TOPMed, which enable the frequency of occurrence of a given allele to be quantified and, coupled with penetrance, this can give a more accurate estimate of the true burden of the disease, by estimating both diagnosed and undiagnosed cases. Exciting developments continue to proceed at pace in this area. For example:
- gnomAD V4.0 was recently released which includes data from 807,162 total individuals, which is nearly five times larger than the combined v2/v3 releases and includes 416,555 individuals from the UK Biobank
- At the recent Festival of Genomics and Biodata, a workshop held by Genomics England, included an update on their coverage of rare diseases in the 100k Genome Project and we will shortly be incorporating this dataset within our program of coverage.
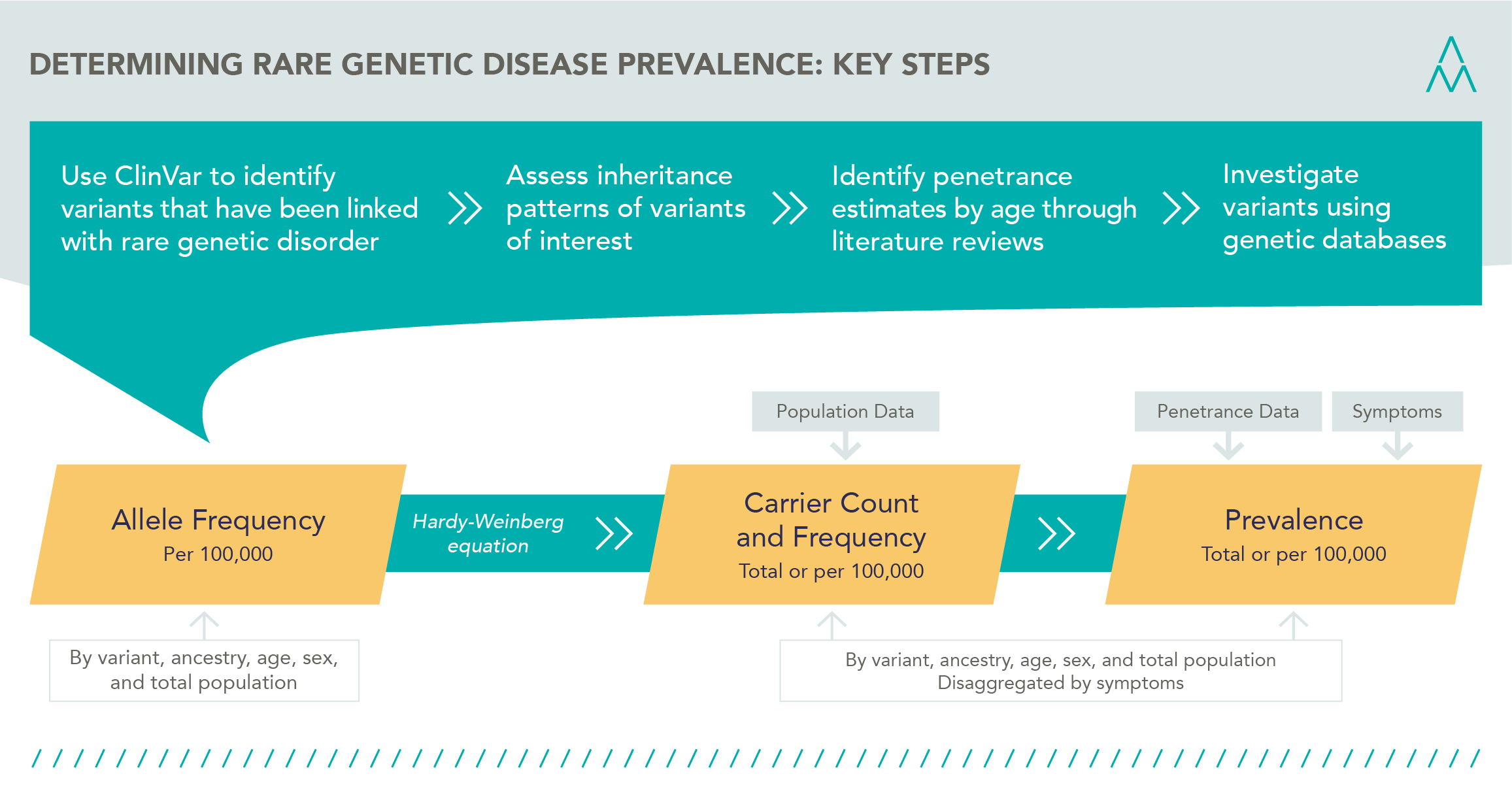
Using the data from these and other databases, our methodology to determine prevalence is illustrated in the graphic below, and includes:
- Determining allele frequency per 100,000, by variant, ancestry, age, sex
- Applying regional or national population data to then estimate allele frequency
- Redistribute and scale to the local ancestry
- Estimating carrier frequency to understand who is at risk of the disease
- Application of penetrance to arrive at prevalence estimates
Senior Evidence Lead Joshua Card-Gowers will be presenting the results of some of our research at the World Orphan Drug Congress in Boston.